Abstract
Introduction: Pediatric mixed phenotype acute leukemia (MPAL), a rare subgroup of leukemia, contains features of both myeloid and lymphoid lineage blasts, which makes the disease more difficult to diagnose/treat. More information is needed to understand the origins of the major pediatric MPAL subtypes, B/Myeloid (B-MPAL) and T/Myeloid (T-MPAL), and how they relate to other leukemias. Single-cell RNA sequencing (scRNA-seq) analysis of bone marrow (BM) can provide in-depth information about the leukemia microenvironment and reveal differences/similarities between MPAL subtypes and other types of leukemia that could be exploited to develop novel diagnostics/therapies.
Methods: We analyzed ~16,000 cells from five pediatric MPAL BM samples to generate a transcriptomic landscape of B-MPAL and T-MPAL blasts and associated microenvironment cells. Samples collected at the time of diagnosis (Dx) were used to generate scRNA-Seq data using a droplet-based barcoding technique (Panigrahy et al. JCI 2019, Tellechea et al. JID, 2020). After data normalization, cell clusters were identified using principal component analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP) approach (Becht et al. Nat. Biotech 2018). Meta-analysis was performed using single cell samples from ongoing studies in the Bhasin lab (Bhasin, et al. Blood 2020 (ASH), Thomas et al. Blood 2020 (ASH)) and publicly available single cell data from GEO biorepository. Unsupervised analysis using UMAP and PCA was performed to determine the overall relationship among B-MPAL, T-MPAL and other leukemias (acute myeloid leukemia (AML), B-cell acute lymphoblastic leukemia (B-ALL), T-cell ALL (T-ALL)). Supervised differentially expressed gene (DEG) analysis was performed to identify B- and T- MPAL blast cell signatures (P value < 0.001 and log2 FC > 0.5). Transcriptomic profiles in MPAL samples and normal BM stem and immune cells were compared using data from the Human Cell Atlas Data Portal (humancellatlas.org). Gene set enrichment analysis (GSEA) was performed, and significantly enriched pathways were compared in MPAL subtypes (P value < 0.001).
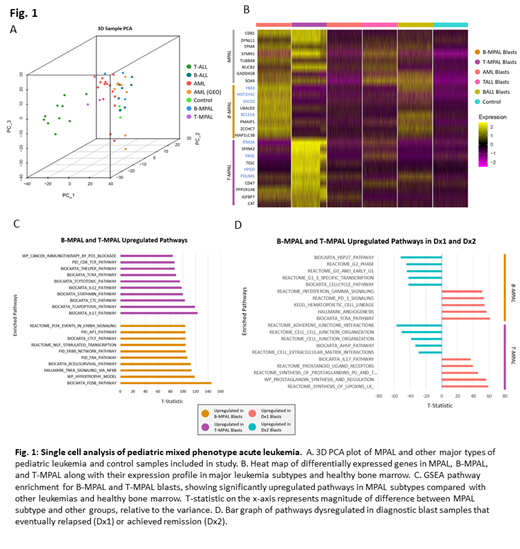
Results: PCA analysis showed transcriptome similarity between B-MPAL and both B-ALL and AML, while T-MPAL transcriptome correlated with T-ALL and AML (Fig. 1A). B- and T-MPAL subtype blasts clustered separately from each other in UMAP analyses, with T-MPAL blasts clustering with T-ALL blasts, and B-MPAL somewhat overlapping with B-ALL blasts. Subtype DEG analysis of leukemia blasts and healthy BM revealed distinct significantly upregulated gene signatures in B-MPAL (YBX3, SOCS2, BCL11A, and HIST1H1C) and T-MPAL (ITM2A, HPGD, PDLIM1, and TRDC) blasts (Fig. 1B). Pathway analysis showed upregulated gene activity related to TNFA signaling via NFKB, B-cell survival, and the AP1, FRA, and NGF transcription factors in B-MPAL blasts. In contrast, IL-17 and IL-12, T-cell apoptosis, and Stathmin pathways were upregulated in T-MPAL blasts (Fig. 1C). T-MPAL T-cells also expressed higher levels of T-cell exhaustion markers compared to T-cells in B-MPAL samples and healthy bone marrow.
After filtering out genes that are significantly expressed in immune cells, we identified genes that are differentially expressed at diagnosis in MPAL blasts from patients that relapsed after treatment (Dx1) versus patients that achieved remission (Dx2). These genes are potential prognostic markers for B-MPAL and T-MPAL relapse/remission. These include MDM2 and NEIL1 from Dx1 and FOSL2 and CDKN1A in Dx2 B-MPAL blasts. In T-MPAL, expression of HES4 and SPINK2 is associated with Dx1 blasts and GNAQ and ITGA4 with Dx2 blasts. Pathway enrichment analysis on B-MPAL blasts revealed upregulation of interferon gamma and PD-1 signaling in Dx1 samples and increased HSP27 and Cell Cycle pathways in the Dx2 subset. T-MPAL Dx1 associated pathways included prostaglandin synthesis and IL-17, while cell-cell junction and extracellular matrix interactions were increased in T-MPAL Dx2 samples (Fig. 1D).
Conclusion: Single-cell profiling was used to characterize the molecular landscapes of MPAL blasts and the bone marrow microenvironment and identified gene signatures and pathways that are specifically enriched in B- and T-MPAL subtypes.
DeRyckere: Meryx: Other: Equity ownership. Graham: Meryx: Membership on an entity's Board of Directors or advisory committees, Other: Equity ownership.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal